(feed readers, this post is better displayed on the web site)
I've been looking for a way to get a structured description of the biographies of the scientists threw the History. One of my investigation led to wikistory, a webstart application based on the data extracted from Wikipedia by the project DBPedia.
However, the data collected from DBPedia are mostly based on the
infoboxes and most of them are missing or are incomplete. (as an example this is ok for
Darwin (
happy birthday) but there is no box for
Georges-Louis Leclerc, Comte de Buffon (
last accessed February 14th 2008 20H46)). Moreover the informations stored in those infoboxes are missing the fields I needed: gender, a short biography, parents, children.... etc....
I eventually decided to go back to look after
freebase wich I
tested a few monthes ago and which was also introduced at
scifoo:
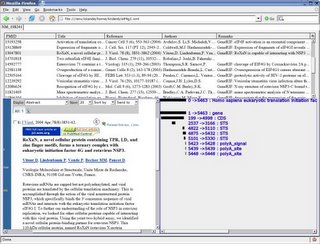
A screenshot of my final result is presented below:
and you can test this interface here:
This is an interactive
XUL page (it will only work with firefox) with a timeline containing a few hundred of scientists.
Just for fun, I also generated a
time-based KML file for google-earth with those data.
The source code used to create this XUL page is available at
here.
Here is how I proceeded:
On freebase I created my own type
"scientist" enclosing some fields such as "short bio", "known for", "students", etc... (I don't know how to define 'inverse properties' in freebase: if A was the teacher of B, how can I automatically say that B was the student of A ?). This type
scientist was added to some of the freebase records and completed (I think that Freebase also parsed the infoboxes in wikipedia to build their database, that is why most their records are almost empty). For example see:
Buffon.
All the persons associated with my type scientist can be retrieved using the following
MQL query:
{"qname1":{"query":[{"guid":null,"type":"/user/lindenb/default_domain/scientist"}]}}
The result looks like this...
{
"status": "200 OK",
"code": "/api/status/ok",
"qname1": {
"code": "/api/status/ok",
"result": [
{
"guid": "#9202a8c04000641f800000000000cb7c",
"type": "/user/lindenb/default_domain/scientist"
},
{
"guid": "#9202a8c04000641f800000000000f65e",
"type": "/user/lindenb/default_domain/scientist"
},
(...)
{
"guid": "#9202a8c04000641f80000000003b7d80",
"type": "/user/lindenb/default_domain/scientist"
},
{
"guid": "#9202a8c04000641f80000000003bd1ef",
"type": "/user/lindenb/default_domain/scientist"
}
]
}
For each
gui we can retrieve the
types associated with the record.
{"qname1":{"query":{"guid":"#9202a8c04000641f800000000000cb7c","type":[]}}}
The
types associated with the record #9202a8c04000641f800000000000cb7c" were returned as follow:
{
"status": "200 OK",
"code": "/api/status/ok",
"qname1": {
"code": "/api/status/ok",
"result": {
"guid": "#9202a8c04000641f800000000000cb7c",
"type": [
"/common/topic",
"/people/person",
"/people/deceased_person",
"/book/author",
"/user/mikelove/default_domain/influence_node",
"/user/lindenb/default_domain/scientist",
"/award/award_winner"
]
}
}
}
For each type, I fetched the fields this record.
{"qname1":{"query":{"guid":"#9202a8c04000641f800000000000cb7c","*":null,"type":"/people/person"}}}
here is the response from freebase:
{
"status": "200 OK",
"code": "/api/status/ok",
"qname1": {
"code": "/api/status/ok",
"result": {
"creator": "/user/metaweb",
"profession": [
"Naturalist",
"Biologist",
"Geologist"
],
"places_lived": [
null,
null,
null,
null
],
"education": [
null,
null
],
"children": [
"George Howard Darwin",
"Horace Darwin"
],
"guid": "#9202a8c04000641f800000000000cb7c",
"employment_history": [],
"id": "/topic/en/charles_darwin",
"religion": [
"Agnosticism",
"Christianity",
"Unitarianism",
"Church of England"
],
"date_of_birth": "
1809-02-12",
"parents": [
"Robert Darwin",
"Susannah Darwin"
],
"metaweb_user_s": [],
"type": "
/people/person",
"attribution": "/user/metaweb",
"permission": "/boot/all_permission",
"timestamp": "2006-10-22T08:53:38.0061Z",
"signature": [],
"weight_kg": null,
"key": [
"Charles_Darwin",
"Charles_Robert_Darwin",
"Darwin$0027s",
"Mary_Darwin",
"Darwin$002C_Charles",
"C$002E_R$002E_Darwin",
"Charles_R$002E_Darwin",
"8145410",
"Charles_Darwin$0027s",
"Charles_darwin",
"charles_darwin",
"CR_Darwin",
"Charles_R_Darwin",
"71b891f5-92bb-42be-9c45-98b8f56a3177"
],
"nationality": [
"United Kingdom"
],
"spouse_s": [
null
],
"name": "Charles Darwin",
"gender": "Male",
"sibling_s": [
null
],
"height_meters": null,
"place_of_birth": "Shrewsbury",
"quotations": []
}
}
}
And so on, using this kind of queries I was able to fetch the birth dates, the geographical coordinate of the places, the pictures, etc...
The result is available here:

http://lindenb.integragen.org/xulhistory/history.php
That's it.
Pierre
Update 2010-08-12 : source code
history.js
var XUL={
NS:"
http://www.mozilla.org/keymaster/gatekeeper/there.is.only.xul"
};
var XHTML={
NS:"
http://www.w3.org/1999/xhtml"
};
function StartDate(year,month,dayOfMonth)
{
this.year=year;
this.month=month;
this.dayOfMonth=dayOfMonth;
}
StartDate.prototype.days=function()
{
var d= this.year*365.25;
if(this.month!=null)
{
d+=(this.month*(365.25/12.0));
if(this.dayOfMonth!=null)
{
d+=this.dayOfMonth;
}
}
return d;
}
function EndDate(year,month,dayOfMonth)
{
this.year=year;
this.month=month;
this.dayOfMonth=dayOfMonth;
}
EndDate.prototype.days=function()
{
var v= 0;
if(this.month!=null)
{
if(this.dayOfMonth!=null)
{
v+=(1+this.dayOfMonth);
v+=this.month*(365.25/12.0);
}
else
{
v+=(this.month+1)*(365.25/12.0);
}
v+=this.year*365.25;
}
else
{
v+=(1+this.year)*365.25;
}
return v;
}
var MY={
now: null,
iconSize:64,
screenWidth:15000,
minDate:null,
maxDate:null,
debug:function(msg)
{
var message=document.getElementById("message");
if(message==null) return;
MY.removeAllChild(message);
message.appendChild(document.createTextNode(msg==null?"null":msg));
},
x1:function(person)
{
return MY.convertDate2Pixel(person.birthDate);
},
x2:function(person)
{
var d= person.deathDate;
if(d==null)
{
d= MY.now;
}
return MY.convertDate2Pixel(d);
},
convertDate2Pixel:function(date)
{
return MY.screenWidth*((date.days()-MY.minDate.days())/(MY.maxDate.days()-MY.minDate.days()));
},
removeAllChild:function(root)
{
if(root==null) return;
while(root.hasChildNodes())
{
root.removeChild(root.firstChild);
}
},
loaded:function()
{
var d= new Date();
MY.now = new EndDate(d.getFullYear(),1+d.getMonth(),d.getUTCDate());
var set= new Array()
for(var i=0;i< persons.length;++i)
{
for(var j=0;j< persons[i].profession.length;++j)
{
set[persons[i].profession[j] ]=1;
}
}
MY.fillListBox("profession",set);
//knownfor
set= new Array()
for(var i=0;i< persons.length;++i)
{
for(var j=0;j< persons[i].knownFor.length;++j)
{
set[persons[i].knownFor[j] ]=1;
}
}
MY.fillListBox("knownfor",set);
//country
set= new Array()
for(var i=0;i< persons.length;++i)
{
for(var j=0;j< persons[i].nationality.length;++j)
{
set[persons[i].nationality[j] ]=1;
}
}
MY.fillListBox("country",set);
//AWARDS
set= new Array()
for(var i=0;i< persons.length;++i)
{
for(var j=0;j< persons[i].awards.length;++j)
{
set[persons[i].awards[j] ]=1;
}
}
MY.fillListBox("awards",set);
MY.pileup();
},
fillListBox:function(id,set)
{
var root=document.getElementById(id);
if(root==null) return;
var array2= new Array(set.length);
for(var p in set)
{
array2.push(p);
}
array2.sort();
for(var j=0;j< array2.length;++j)
{
var item=document.createElementNS(XUL.NS,"listitem");
item.setAttribute("label",array2[j]);
item.setAttribute("value",array2[j]);
root.appendChild(item);
}
},
selectedItems:function(id)
{
var set= new Array();
var root=document.getElementById(id);
if(root==null) return set;
var selected= root.selectedItems;
for(var i=0;i<selected.length;++i)
{
set.push(selected[i].value);
}
return set;
},
date2text:function(date)
{
var s=""+date.year;
if(date.month!=null)
{
s+=" ";
switch(date.month)
{
case 1: s+=("Jan"); break;
case 2: s+=("Feb"); break;
case 3: s+=("Mar"); break;
case 4: s+=("Apr"); break;
case 5: s+=("May"); break;
case 6: s+=("Jun"); break;
case 7: s+=("Jul"); break;
case 8: s+=("Aug"); break;
case 9: s+=("Sep"); break;
case 10: s+=("Oct"); break;
case 11: s+=("Nov"); break;
case 12: s+=("Dec"); break;
default: s+=date.month; break;
}
if(date.dayOfMonth!=null)
{
s+=" "+date.dayOfMonth;
}
}
return s;
},
containsSet:function(set,subset)
{
if(subset.length==0) return true;
for(var i=0;i< subset.length;++i)
{
if(set.indexOf(subset[i])!=-1) return true;
}
return false;
},
update:function()
{
for(var i=0;i< persons.length;++i)
{
persons[i].selected=true;
}
var sel=MY.selectedItems("profession");
for(var i=0;i< persons.length && sel.length>0;++i)
{
if(!MY.containsSet(persons[i].profession,sel))
{
persons[i].selected=false;
}
}
sel=MY.selectedItems("knownfor");
for(var i=0;i< persons.length && sel.length>0;++i)
{
if(!persons[i].selected) continue;
if(!MY.containsSet(persons[i].knownFor,sel))
{
persons[i].selected=false;
}
}
sel=MY.selectedItems("country");
for(var i=0;i< persons.length && sel.length>0;++i)
{
if(!persons[i].selected) continue;
if(!MY.containsSet(persons[i].nationality,sel))
{
persons[i].selected=false;
}
}
sel=MY.selectedItems("awards");
for(var i=0;i< persons.length && sel.length>0;++i)
{
if(!persons[i].selected) continue;
if(!MY.containsSet(persons[i].awards,sel))
{
persons[i].selected=false;
}
}
sel=MY.selectedItems("gender");
for(var i=0;i< persons.length && sel.length>0;++i)
{
if(!persons[i].selected ) continue;
if(sel.indexOf(persons[i].gender)==-1)
{
persons[i].selected=false;
}
}
for(var i=0;i< persons.length ;++i)
{
persons[i].node.style.opacity=(persons[i].selected?1.0:0.3);
}
},
simpleTag:function(tag,text)
{
var e = document.createElementNS(XHTML.NS,tag);
e.appendChild(document.createTextNode(text));
return e;
},
bold:function(text) { return MY.simpleTag("h:b",text);},
italic:function(text) { return MY.simpleTag("h:i",text);},
underline:function(text) { return MY.simpleTag("h:u",text);},
pileup:function()
{
var remains=new Array(persons.length);
for(var i=0;i< persons.length;++i) remains[i]=persons[i];
MY.minDate=null;
MY.maxDate=null;
for(var i=0;i< remains.length;++i)
{
var o=remains[i];
if(MY.minDate==null || o.birthDate.days() < MY.minDate.days())
{
MY.minDate= o.birthDate;
}
if(o.deathDate!=null && (MY.maxDate==null || MY.maxDate.days()< o.deathDate.days()))
{
MY.maxDate= o.deathDate;
}
}
if(MY.minDate==null || MY.maxDate==null) return;
/*
var nLine=-1;
while(remains.length>0)
{
++nLine;
var first=remains[0];
remains=remains.slice(1);
first.y=nLine;
while(true)
{
var best=null;
var bestIndex=-1;
for(var i=0;i< remains.length;++i)
{
var next=remains[i];
if(MY.x1(next)< MY.x2(first)+5) continue;
if(best==null ||
(MY.x1(next)-MY.x2(first) < MY.x1(best)-MY.x2(first)))
{
best=next;
bestIndex=i;
}
}
if(best==null) break;
first=best;
first.y=nLine;
remains.splice(bestIndex,1);
}
}
*/
var timeline=document.getElementById("timeline");
MY.removeAllChild(timeline);
var MARGIN=2;
var HEIGHT=MY.iconSize+MARGIN*2;
for(var i=0;i< persons.length;++i)
{
var o= persons[i];
var stack= document.createElementNS(XUL.NS,"stack");
var style="top:"+Math.round(o.y*(HEIGHT+10))+"px;"+
"left:"+Math.round(o.x1)+"px;"+
"width:"+Math.round(o.x2-o.x1)+"px;"+
"height:"+Math.round(HEIGHT)+"px;"+
"background-color:black;"+
"color:white;"+
"border-width:2px;"+
"border-color:red;"+
"overflow:hidden;"+
"font-size:11px;"+
"opacity: 1;"
;
o.node=stack;
stack.setAttribute("style",style);
var hbox= document.createElementNS(XUL.NS,"hbox");
hbox.setAttribute("flex","1");
stack.appendChild(hbox);
if(o.img!=null)
{
var img= document.createElementNS(XUL.NS,"image");
hbox.appendChild(img);
img.setAttribute("src",o.img);
img.setAttribute("style","width:"+MY.iconSize+"px;height:"+MY.iconSize+"px;");
}
var div= document.createElementNS(XHTML.NS,"h:div");
div.setAttribute("style","width:"+Math.round(MY.x2(o)-MY.x1(o)-(o.img==null?0:MY.iconSize))+"px;");
//div= document.createElementNS(XUL.NS,"label");
hbox.appendChild(div);
var anchor= document.createElementNS(XHTML.NS,"h:a");
anchor.appendChild(document.createTextNode(o.name));
anchor.setAttribute("href","
http://www.freebase.com/view/guid/"+o.guid);
anchor.setAttribute("target",o.guid);
anchor.setAttribute("title",o.name);
div.appendChild(anchor);
div.appendChild(document.createTextNode(" : "));
if(o.birthDate!=null)
{
div.appendChild(document.createTextNode(MY.date2text(o.birthDate)));
if(o.birthPlace!=null)
{
div.appendChild(document.createTextNode(" at "+o.birthPlace));
}
div.appendChild(document.createTextNode(" - "));
}
if(o.deathDate!=null)
{
div.appendChild(document.createTextNode(MY.date2text(o.deathDate)));
if(o.deathPlace!=null)
{
div.appendChild(document.createTextNode(" at "+o.deathPlace));
}
}
div.appendChild(document.createTextNode(":"+o.shortBio+" "));
if(o.knownFor.length>0)
{
div.appendChild(MY.bold("Known For:"));
for(var k in o.knownFor)
{
div.appendChild(document.createTextNode(o.knownFor[k]+" "));
}
}
timeline.appendChild(stack);
}
}
};
history.xul
<?xml version="1.0" encoding="UTF-8"?>
<?xml-stylesheet href="chrome://global/skin/" type="text/css"?>
<window
id="main-window"
title="History Of Science"
orient="horizontal"
xmlns="
http://www.mozilla.org/keymaster/gatekeeper/there.is.only.xul"
xmlns:h="
http://www.w3.org/1999/xhtml"
onload="MY.loaded();"
>
<!-- FIREFOX 2.0 IS REQUIRED TO SEE THIS FILE !!! -->
<script src="history.js"/>
<script src="person.js"/>
<script src="
http://www.google-analytics.com/urchin.js" type="text/javascript"></script><script type="text/javascript">_uacct = "UA-307413-2";urchinTracker();</script>
<vbox flex="1">
<hbox>
<h:div><h:h1>History Of Sciences</h:h1> 2008 <h:a title="plindenbaum@yahoo.fr" href="
mailto:plindenbaum@yahoo.fr">Pierre Lindenbaum PhD</h:a>.
Testing <h:a title="freebase" href="
http://www.freebase.com">Freebase</h:a> to fetch the biographies of famous scientists. See also my <h:a href="
http://plindenbaum.blogspot.com/2008/02/freebase-and-history-of-sciences.html">blog</h:a> and the <h:a href="
http://maps.google.com/maps?f=q&hl=en&geocode=&q=http%3A%2F%2Flindenb.integragen.org%2Fxulhistory%2Fhistory.kml&ie=UTF8&ll=53.956086,-13.007812&spn=99.233765,284.0625&t=h&z=2" title="kml">KML file</h:a> for Google Earth. </h:div>
</hbox>
<scrollbox flex="6" style='overflow: auto;'>
<stack id="timeline" flex="6" style="background-color:lightgray;color:black;width:200px;border-style:solid;border-color: black;border-width:1px;">
<!-- content goes here -->
</stack>
</scrollbox>
<vbox flex="1">
<hbox><description id="message"/></hbox>
<hbox flex="1">
<listbox id="gender" seltype="multiple" rows="2" flex="1">
<listhead>
<listheader label="Gender"/>
</listhead>
<listitem label="Man" value="Man"/>
<listitem label="Woman" value="Female"/>
</listbox>
<listbox id="profession" seltype="multiple" rows="5" flex="1">
<listhead>
<listheader label="Profession"/>
</listhead>
</listbox>
<listbox id="country" seltype="multiple" rows="5" flex="1">
<listhead>
<listheader label="Country"/>
</listhead>
</listbox>
<listbox id="awards" seltype="multiple" rows="5" flex="1">
<listhead>
<listheader label="Awards"/>
</listhead>
</listbox>
<listbox id="knownfor" seltype="multiple" rows="5" flex="1">
<listhead>
<listheader label="Known for"/>
</listhead>
</listbox>
<button id="yes" label="Update" flex="1" oncommand="MY.update();"/>
</hbox>
<hbox><label value="Updated: 2008-03-02"/></hbox>
</vbox>
</vbox>
</window>
person.js
var persons=[
{
name:"Thales",
guid:"9202a8c04000641f800000000003b246",
gender:"Male",
x1:0.0,
x2:527.9741361723788,
y:0,
node:null,
selected:true,
nationality:["Ancient Greece"],
shortBio:"pre-Socratic Greek philosopher and one of the Seven Sages of Greece. Many regard him as the first philosopher in the Greek tradition, while some also consider him the \"father of science.\"",
profession:["Mathematician","Philosopher"],
birthDate:new StartDate(-634,null,null),
birthPlace:null,deathDate:new EndDate(-542,null,null),
deathPlace:null,
knownFor:["Thales\' theorem"],
img:null,
awards:[]
}
,
{
name:"Anaximander",
guid:"9202a8c04000641f8000000000004e68",
gender:"Male",
x1:147.60567247829948,
x2:505.2655711757174,
y:1,
node:null,
selected:true,
nationality:["Ancient Greece"],
shortBio:"pre-Socratic philosopher",
profession:["Mathematician"],
birthDate:new StartDate(-608,null,null),
birthPlace:null,deathDate:new EndDate(-546,null,null),
deathPlace:null,
knownFor:["Apeiron"],
img:null,
awards:[]
}
(...)
,
{
name:"Brian Greene",
guid:"9202a8c04000641f80000000001f50ef",
gender:"Male",
x1:14744.621902781384,
x2:15000.473095104098,
y:5,
node:null,
selected:true,
nationality:["United States"],
shortBio:"Theoretical physicist and one of the best-known string theorists.",
profession:["Physicist","Scientist","Science writer"],
birthDate:new StartDate(1963,2,9),
birthPlace:"New York, New York",deathDate:null,
deathPlace:null,
knownFor:["The Elegant Universe","The Fabric of the Cosmos","String theory"],
img:"9202a8c04000641f80000000049ae0ba.png",
awards:[]
}
];