20 December 2013
Publié par
Pierre Lindenbaum
at
7:10 PM
0
commentaires
![]()
Libellés : bioinformatics, make, pipeline, workflow
12 December 2013
Inside Jvarkit: view BAM, cut, stats, head, tail, shuffle, downsample, group-by-gene VCFs...
Here are a few tools I recently wrote (and reinvented) for Jvarkit.
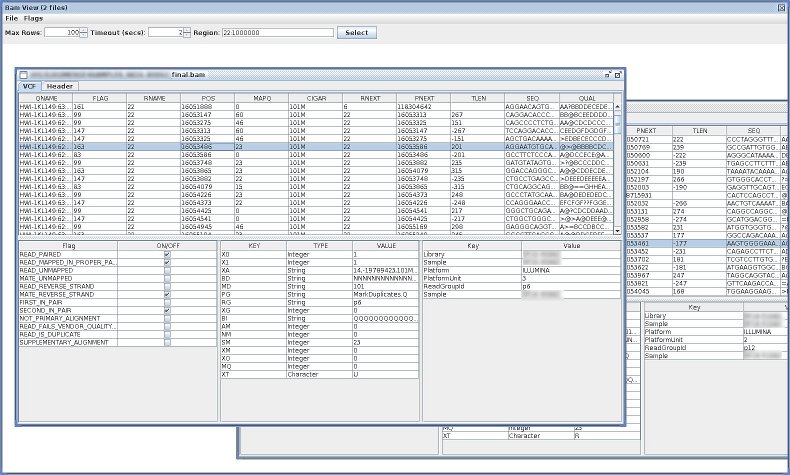
- BamViewGui
- a simple java-Swing-based BAM viewer.
- VcfShuffle
- Shuffle a VCF.
- GroupByGene
- Group VCF data by Gene
$ curl -s -k "https://raw.github.com/arq5x/gemini/master/test/test4.vep.snpeff.vcf" |\ java -jar dist/groupbygene.jar |\ head | column -t #chrom min.POS max.POS gene.name gene.type samples.affected count.variations M10475 M10478 M10500 M128215 chr10 52004315 52004315 ASAH2 snpeff-gene-name 2 1 0 0 1 1 chr10 52004315 52004315 ASAH2 vep-gene-name 2 1 0 0 1 1 chr10 52497529 52497529 ASAH2B snpeff-gene-name 2 1 0 1 1 0 chr10 52497529 52497529 ASAH2B vep-gene-name 2 1 0 1 1 0 chr10 48003992 48003992 ASAH2C snpeff-gene-name 3 1 1 1 1 0 chr10 48003992 48003992 ASAH2C vep-gene-name 3 1 1 1 1 0 chr10 126678092 126678092 CTBP2 snpeff-gene-name 1 1 0 0 0 1 chr10 126678092 126678092 CTBP2 vep-gene-name 1 1 0 0 0 1 chr10 135336656 135369532 CYP2E1 snpeff-gene-name 3 2 0 2 1 1
- DownSampleVcf
- Down sample a VCF.
- VcfHead
- Print the first variants of a VCF.
- VcfTail
- Print the last variants of a VCF
- VcfCutSamples
- Select/Exclude some samples from a VCF
- VcfStats>
- Generate some statistics from a VCF. The ouput is a XML file that can be processed with xslt.
$ curl "https://raw.github.com/arq5x/gemini/master/test/test4.vep.snpeff.vcf" |\
java -jar dist/vcfstats.jar |\
xmllint --format -
<?xml version="1.0" encoding="UTF-8"?>
<vcf-statistics version="314bf88924a4003e6d6189ad3280d8b4df485aa1" input="stdin" date="Thu Dec 12 16:20:14 CET 2013">
<section name="General">
<statistics name="general" description="general">
<counts name="general" description="General" keytype="string">
<property key="num.dictionary.chromosomes">93<
(...)
That's it,
Pierre
Publié par
Pierre Lindenbaum
at
4:54 PM
2
commentaires
![]()
Libellés : bioinformatics, java, ngs
20 November 2013
Inside Jvarkit: Shrink your fastqs by 40% by aligning them to REF before compression.
BamToFastq is an implementation of https://twitter.com/DNAntonie/status/402909852277932032"
Shrink your FASTQ.bz2 files by 40+% using this one weird tip -> order them by alignment to reference before compression!
— Antonie DNA Software (@DNAntonie) November 19, 2013Example : piping bwa mem
$ bwa mem -M human_g1k_v37.fasta Sample1_L001_R1_001.fastq.gz Sample2_S5_L001_R2_001.fastq.gz |\ java -jar dist/bam2fastq.jar -F tmpR1.fastq.gz -R tmpR2.fastq.gz
$ ls -lah Sample1_L001_R1_001.fastq.gz Sample2_S5_L001_R2_001.fastq.gz -rw-r--r-- 1 lindenb lindenb 181M Jun 14 15:20 Sample1_L001_R1_001.fastq.gz -rw-r--r-- 1 lindenb lindenb 190M Jun 14 15:20 Sample1_L001_R2_001.fastq.gz
after (these are Haloplex Data, with a lot of duplicates )
$ ls -lah tmpR1.fastq.gz tmpR2.fastq.gz -rw-rw-r-- 1 lindenb lindenb 96M Nov 20 17:10 tmpR1.fastq.gz -rw-rw-r-- 1 lindenb lindenb 106M Nov 20 17:10 tmpR2.fastq.gz
using BZ2:
$ ls -lah *.bz2 -rw-rw-r-- 1 lindenb lindenb 77M Nov 20 17:55 tmpR1.fastq.bz2 -rw-rw-r-- 1 lindenb lindenb 87M Nov 20 17:55 tmpR2.fastq.bz2
That's it
Pierre
Publié par
Pierre Lindenbaum
at
6:55 PM
2
commentaires
![]()
Libellés : bioinformatics, java, ngs
30 October 2013
GNU Make: saving the versions of the tools using 'order-only-prerequisites' : my notebook
Rule 3 of "Ten Simple Rules for Reproducible Computational Research". is
:
I work with Makefile-based workflows: how can I save the version of each software used when I invoke 'make', whatever the target is ? A naive solution is to add a dependency to each target. For example, the following makefile takes a simple SAM file, convert it to BAM, sort and index. For each target, I've added a dependency named "dump_params" that append the version of samtools to a file
"config.txt".But that solution doesn't work because make re-builds all targets even if the top target already exists.
$ make date << config.txt && \ echo -n "Samtools " << config.txt && \ samtools 2<&1 | grep Version << config.txt samtools view -Sb samtools-0.1.18/examples/toy.sam < unsorted.bam [samopen] SAM header is present: 2 sequences. samtools sort unsorted.bam sorted samtools index sorted.bam $ make date << config.txt && \ echo -n "Samtools " << config.txt && \ samtools 2<&1 | grep Version << config.txt samtools view -Sb samtools-0.1.18/examples/toy.sam < unsorted.bam [samopen] SAM header is present: 2 sequences. samtools sort unsorted.bam sorted samtools index sorted.bam
The solution I got via Stackoverflow is to use a order-only-prerequisites: "Order-only prerequisites can be specified by placing a pipe symbol (|) in the prerequisites list: any prerequisites to the left of the pipe symbol are normal; any prerequisites to the right are order-only... (...) Note that if you declare the same file to be both a normal and an order-only prerequisite, the normal prerequisite takes precedence (...)". The makefile with the 'order-only-prerequisites' is now:
And that works ! the final target is generated only once, but the file 'config.txt' is always generated.
$ make date << config.txt && \ echo -n "Samtools " << config.txt && \ samtools 2<&1 | grep Version << config.txt samtools view -Sb samtools-0.1.18/examples/toy.sam < unsorted.bam [samopen] SAM header is present: 2 sequences. samtools sort unsorted.bam sorted samtools index sorted.bam $ make date << config.txt && \ echo -n "Samtools " << config.txt && \ samtools 2<&1 | grep Version << config.txt $ make date << config.txt && \ echo -n "Samtools " << config.txt && \ samtools 2<&1 | grep Version << config.txtThat's it,
Pierre
Update :another solution
Citing MadScientist's answer on stackoverflow : Another option is to use immediately expanded shell functions, like:__dummy := $(shell echo "Makefile was run." >> config.txt)Since it's immediately expanded the shell script will be invoked once, as the makefile is read in. There's no need to define a dump_params target or include it as a prerequisite. This is more old-school, but has the advantage that it will run for every invocation of make, without having to go through and ensure every target has the proper order-only prerequisite defined.
Publié par
Pierre Lindenbaum
at
6:52 PM
1 commentaires
![]()
Libellés : bioinformatics, make, pipeline, samtools, workflow
25 October 2013
YES: "Choice of transcripts and software has a large effect on variant annotation"
This post was inspired by Aaron Quinlan's tweet:
Poster 1485: variant annotation is harder than you think. transcript set matters (surprise). #ASHG2013 http://t.co/FgJzomqVuV
— Aaron Quinlan (@aaronquinlan) October 23, 2013
Here is an example of a missense mutation found with VCFPredictions, a simple tool I wrote for variant effect prediction.
#CHROM POS ID ALT REF 1 23710805 rs605468 A Gmy tool uses the UCSC knownGene track, here is the context of the variant in the UCSC genome browser. There is one exon for TCEA3 (uc021oig.1) on the reverse strand.
If the base at 23710805 is changed from 'A'→'G' on the forward strand, there will be a non-synonymous variation Y(TAT)→H(CAT) on the reverse strand.
At the NCBI rs605468 is said to be " located in the intron region of NM_003196.1."
VEP cannot find this missense variation:
Uploaded Variation Location Allele Gene Feature Feature type Consequence Position in cDNA Position in CDS Position in protein Amino acid change Codon change Co-located Variation Extra rs605468 1:23710805 G - ENSR00001518296 Transcript regulatory_region_variant - - - - - rs605468 GMAF=A:0.1204 rs605468 1:23710805 G ENSG00000204219 ENST00000476978 Transcript intron_variant - - - - - rs605468 GMAF=A:0.1204 rs605468 1:23710805 G ENSG00000204219 ENST00000450454 Transcript intron_variant - - - - - rs605468 GMAF=A:0.1204
(of course, my tool doesn't find some variations found in VEP too)
That's it,
Pierre
Publié par
Pierre Lindenbaum
at
11:10 AM
0
commentaires
![]()
Libellés : bioinformatics, ncbi, variation, vcf
24 October 2013
PubMed Commons & Bioinformatics: a call for action
NCBI pubmed Commons/@PubMedCommons is a new system that enables researchers to share their opinions about scientific publications. Researchers can comment on any publication indexed by PubMed, and read the comments of others.
Now that we can add some comments to the papers in pubmed, I suggest to flag the articles to mark the deprecated softwares, databases, hyperlinks using a simple controlled syntax. Here are a few examples: the line starts with '!MOVED' or '!NOTFOUND' and is followed by a URL or/and a list of PMID or/and a quoted comment.
Examples
!MOVED: for http://www.ncbi.nlm.nih.gov/pubmed/8392714 (Rebase/1993) to http://www.ncbi.nlm.nih.gov/pubmed/19846593 (Rebase/2010)!MOVED PMID:19846593 "A more recent version"In http://www.ncbi.nlm.nih.gov/pubmed/19919682 the URL has moved to http://biogps.org.
!MOVED <http://biogps.org>I moved the sources of http://www.ncbi.nlm.nih.gov/pubmed/9682060 to github
!MOVED <https://github.com/lindenb/cloneit/tree/master/c>!NOTFOUND: for http://www.ncbi.nlm.nih.gov/pubmed/9545460 ( Biocat EXCEL template ) url http://www.ebi.ac.uk/biocat/biocat.html returns a 404.
!NOTFOUND "The URL http://www.ebi.ac.uk/biocat/biocat.html was not found "
That's it,
Pierre
Publié par
Pierre Lindenbaum
at
1:06 PM
1 commentaires
![]()
23 October 2013
Inside the variation toolkit: Generating a structured document describing an Illumina directory.
I wrote a tool named "Illuminadir" : it creates a structured (JSON or XML) representation of a directory containing some Illumina FASTQs (I only tested it with HiSeq , paired end-data and indexes).
Motivation
Illuminadir scans folders , search for FASTQs and generate a structured summary of the files (xml or json).
Currently only tested with HiSeq data having an index
Compilation
See also Compilation
ant illuminadir
Options
| Option | Description |
|---|---|
| IN=File | root directories This option may be specified 1 or more times. |
| JSON=Boolean | json output Default value: false. Ouput, could be used with jsvelocity https://github.com/lindenb/jsvelocity |
Example
$ java -jar dist/illuminadir.jar \ I=dir1 \ I=dir2 | xsltproc xml2script.xslt > script.bash (...)
XML output
The XML ouput looks like this:
<?xml version="1.0" encoding="UTF-8"?> <illumina> <!--com.github.lindenb.jvarkit.tools.misc.IlluminaDirectory IN=[RUN62_XFC2DM8ACXX/data] JSON=false VERBOSITY=INFO QUIET=false VALIDATION_STRINGENCY=STRICT COMPRESSION_LEVEL=5 MAX_RECORDS_IN_RAM=500000 CREATE_INDEX=false CREATE_MD5_FILE=false--> <directory path="RUN62_XFC2DM8ACXX/data"> <samples> <sample name="SAMPLE1"> <pair md5="cd4b436ce7aff4cf669d282c6d9a7899" lane="8" index="ATCACG" split="2"> <fastq md5filename="3369c3457d6603f06379b654cb78e696" side="1" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_002.fastq.gz" file-size="359046311"/> <fastq md5filename="832039fa00b5f40108848e48eb437e0b" side="2" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R2_002.fastq.gz" file-size="359659451"/> </pair> <pair md5="b3050fa3307e63ab9790b0e263c5d240" lane="8" index="ATCACG" split="3"> <fastq md5filename="091727bb6b300e463c3d708e157436ab" side="1" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_003.fastq.gz" file-size="206660736"/> <fastq md5filename="20235ef4ec8845515beb4e13da34b5d3" side="2" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R2_003.fastq.gz" file-size="206715143"/> </pair> <pair md5="9f7ee49e87d01610372c43ab928939f6" lane="8" index="ATCACG" split="1"> <fastq md5filename="54cb2fd33edd5c2e787287ccf1595952" side="1" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_001.fastq.gz" file-size="354530831"/> <fastq md5filename="e937cbdf32020074e50d3332c67cf6b3" side="2" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R2_001.fastq.gz" file-size="356908963"/> </pair> <pair md5="0697846a504158eef523c0f4ede85288" lane="7" index="ATCACG" split="2"> <fastq md5filename="6fb35d130efae4dcfa79260281504aa3" side="1" path="RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L007_R1_002.fastq.gz" file-size="357120615"/> (...) <pair md5="634cbb29ca64604174963a4fff09f37a" lane="7" split="1"> <fastq md5filename="bc0b283a58946fd75a95b330e0aefdc8" side="1" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane7/lane7_Undetermined_L007_R1_001.fastq.gz" file-size="371063045"/> <fastq md5filename="9eab26c5b593d50d642399d172a11835" side="2" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane7/lane7_Undetermined_L007_R2_001.fastq.gz" file-size="372221753"/> </pair> <pair md5="bf31099075d6c3c7ea052b8038cb4a03" lane="8" split="2"> <fastq md5filename="f229389da36a3efc20888bffdec09b80" side="1" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane8/lane8_Undetermined_L008_R1_002.fastq.gz" file-size="374331268"/> <fastq md5filename="417fd9f28d24f63ce0d0808d97543315" side="2" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane8/lane8_Undetermined_L008_R2_002.fastq.gz" file-size="372181102"/> </pair> <pair md5="95cab850b0608c53e8c83b25cfdb3b2b" lane="8" split="3"> <fastq md5filename="23f5be8a962697f50e2a271394242e2f" side="1" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane8/lane8_Undetermined_L008_R1_003.fastq.gz" file-size="60303589"/> <fastq md5filename="3f39f212c36d0aa884b81649ad56630c" side="2" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane8/lane8_Undetermined_L008_R2_003.fastq.gz" file-size="59123627"/> </pair> <pair md5="ab108b1dda7df86f33f375367b86bfe4" lane="8" split="1"> <fastq md5filename="14f8281cf7d1a53d29cd03cb53a45b4a" side="1" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane8/lane8_Undetermined_L008_R1_001.fastq.gz" file-size="371255111"/> <fastq md5filename="977fd388e1b3451dfcdbf9bdcbb89ed4" side="2" path="RUN62_XFC2DM8ACXX/data/Undetermined_indices/Sample_lane8/lane8_Undetermined_L008_R2_001.fastq.gz" file-size="370744530"/> </pair> </undetermined> </directory> </illumina>
How to use that file ? here is a example of XSLT stylesheet that can generate a Makefile to generate a LaTeX about the number of reads per Lane/Sample/Index
<?xml version='1.0' encoding="ISO-8859-1"?> <xsl:stylesheet xmlns:xsl='http://www.w3.org/1999/XSL/Transform' version='1.0' > <xsl:output method="text"/> <xsl:template match="/"> .PHONY:all clean all: report.pdf report.pdf: report.tex pdflatex $< report.tex : all.count echo 'T<-read.table("$<",head=TRUE,sep="\t");$(foreach FTYPE,Index Sample Lane, T2<-tapply(T$$count,T$$${FTYPE},sum);png("${FTYPE}.png");barplot(T2,las=3);dev.off();)' | R --no-save echo "\documentclass{report}" > $@ echo "\usepackage{graphicx}" >> $@ echo "\date{\today}" >> $@ echo "\title{FastQ Report}" >> $@ echo "\begin{document}" >> $@ echo "\maketitle" >> $@ $(foreach FTYPE,Index Sample Lane, echo "\section{By ${FTYPE}}#\begin{center}#\includegraphics{${FTYPE}.png}#\end{center}" | tr "#" "\n" >> $@ ; ) echo "\end{document}" >> $@ all.count : $(addsuffix .count, <xsl:for-each select="//fastq" ><xsl:value-of select="@md5filename"/><xsl:text> </xsl:text></xsl:for-each>) echo -e "Lane\tsplit\tside\tsize\tcount\tIndex\tSample" > $@ && \ cat $^ >> $@ <xsl:apply-templates select="//fastq" mode="count"/> clean: rm -f all.count report.pdf report.tex $(addsuffix .count, <xsl:for-each select="//fastq" ><xsl:value-of select="@md5filename"/><xsl:text> </xsl:text></xsl:for-each>) </xsl:template> <xsl:template match="fastq" mode="count"> $(addsuffix .count, <xsl:value-of select="@md5filename"/>): <xsl:value-of select="@path"/> gunzip -c $< | awk '(NR%4==1)' | wc -l | xargs printf "<xsl:value-of select="../@lane"/>\t<xsl:value-of select="../@split"/>\t<xsl:value-of select="@side"/>\t<xsl:value-of select="@file-size"/>\t%s\t<xsl:choose><xsl:when test="../@index"><xsl:value-of select="../@index"/></xsl:when><xsl:otherwise>Undetermined</xsl:otherwise></xsl:choose>\t<xsl:choose><xsl:when test="../../@name"><xsl:value-of select="../../@name"/></xsl:when><xsl:otherwise>Undetermined</xsl:otherwise></xsl:choose>\n" > $@ </xsl:template> </xsl:stylesheet>
$ xsltproc illumina.xml illumina2makefile.xsl > Makefile
.PHONY:all clean all: report.pdf report.pdf: report.tex pdflatex $< report.tex : all.count echo 'T<-read.table("$<",head=TRUE,sep="\t");$(foreach FTYPE,Index Sample Lane, T2<-tapply(T$$count,T$$${FTYPE},sum);png("${FTYPE}.png");barplot(T2,las=3);dev.off();)' | R --no-save echo "\documentclass{report}" > $@ echo "\usepackage{graphicx}" >> $@ echo "\date{\today}" >> $@ echo "\title{FastQ Report}" >> $@ echo "\begin{document}" >> $@ echo "\maketitle" >> $@ $(foreach FTYPE,Index Sample Lane, echo "\section{By ${FTYPE}}#\begin{center}#\includegraphics{${FTYPE}.png}#\end{center}" | tr "#" "\n" >> $@ ; ) echo "\end{document}" >> $@ all.count : $(addsuffix .count, 3369c3457d6603f06379b654cb78e696 832039fa00b5f40108848e48eb437e0b 091727bb6b300e463c3d708e157436ab 20235ef4ec88....) echo -e "Lane\tsplit\tside\tsize\tcount\tIndex\tSample" > $@ && \ cat $^ >> $@ $(addsuffix .count, 3369c3457d6603f06379b654cb78e696): RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_002.fastq.gz gunzip -c $< | awk '(NR%4==1)' | wc -l | xargs printf "8\t2\t1\t359046311\t%s\tATCACG\tSAMPLE1\n" > $@ $(addsuffix .count, 832039fa00b5f40108848e48eb437e0b): RUN62_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R2_002.fastq.gz gunzip -c $< | awk '(NR%4==1)' | wc -l | xargs printf "8\t2\t2\t359659451\t%s\tATCACG\tSAMPLE1\n" > $@ (....)
JSON output
The JSON output looks like this
[{"directory":"RUN62_XFC2DM8ACXX/data","samples":[{"sample":"SAMPLE1","files":[{ "md5pair":"cd4b436ce7aff4cf669d282c6d9a7899","lane":8,"index":"ATCACG","split":2 ,"forward":{"md5filename":"3369c3457d6603f06379b654cb78e696","path":"20131001_SN L149_0062_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_002.fastq.g z","side":1,"file-size":359046311},"reverse":{"md5filename":"832039fa00b5f401088 48e48eb437e0b","path":"20131001_SNL149_0062_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/ SAMPLE1_ATCACG_L008_R2_002.fastq.gz","side":2,"file-size":359659451}},{"md5pair" :"b3050fa3307e63ab9790b0e263c5d240","lane":8,"index":"ATCACG","split":3,"forward ":{"md5filename":"091727bb6b300e463c3d708e157436ab","path":"20131001_SNL149_0062 _XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_003.fastq.gz","side" :1,"file-size":206660736},"reverse":{"md5filename":"20235ef4ec8845515beb4e13da34 b5d3","path":"20131001_SNL149_0062_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_A TCACG_L008_R2_003.fastq.gz","side":2,"file-size":206715143}},{"md5pair":"9f7ee49 e87d01610372c43ab928939f6","lane":8,"index":"ATCACG","split":1,"forward":{"md5fi lename":"54cb2fd33edd5c2e787287ccf1595952","path":"20131001_SNL149_0062_XFC2DM8A CXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L008_R1_001.fastq.gz","side":1,"file- size":354530831},"reverse":{"md5filename":"e937cbdf32020074e50d3332c67cf6b3","pa th":"20131001_SNL149_0062_XFC2DM8ACXX/data/OUT/Sample_SAMPLE1/SAMPLE1_ATCACG_L00 8_R2_001.fastq.gz","side":2,"file-size":356908963}},{"md5pair":"0697846a504158ee f523c0f4ede85288","lane":7,"index":"ATCACG","split":2,"forward":{"md5filename":"
The velocity template for jsvelocity
#macro(maketarget $fastq) $(addsuffix .count, ${fastq.md5filename}): ${fastq.path} gunzip -c $< | awk '(NR%4==1)' | wc -l | xargs printf "${fastq.parentNode.lane}\t${fastq.parentNode.split}\t${fastq.side}\t${fastq['file-size']}\t%s\t#if(${fastq.parentNode.containsKey("index")})${fastq.parentNode.index}#{else}Undetermined#{end}\t#if(${fastq.parentNode.parentNode.containsKey("name")})${fastq.parentNode.parentNode.name}#{else}Undetermined#{end}\n" > $@ #end .PHONY:all clean all: report.pdf report.pdf: report.tex pdflatex $< report.tex : all.count echo 'T<-read.table("$<",head=TRUE,sep="\t");$(foreach FTYPE,Index Sample Lane, T2<-tapply(T$$count,T$$${FTYPE},sum);png("${FTYPE}.png");barplot(T2,las=3);dev.off();)' | R --no-save echo "\documentclass{report}" > $@ echo "\usepackage{graphicx}" >> $@ echo "\date{\today}" >> $@ echo "\title{FastQ Report}" >> $@ echo "\begin{document}" >> $@ echo "\maketitle" >> $@ $(foreach FTYPE,Index Sample Lane, echo "\section{By ${FTYPE}}#\begin{center}#\includegraphics{${FTYPE}.png}#\end{center}" | tr "#" "\n" >> $@ ; ) echo "\end{document}" >> $@ all.count : $(addsuffix .count, #foreach($dir in $all) #foreach($sample in ${dir.samples})#foreach($pair in ${sample.files}) ${pair.forward.md5filename} ${pair.reverse.md5filename} #end #end #foreach($pair in ${dir.undetermined}) ${pair.forward.md5filename} ${pair.reverse.md5filename} #end #end ) #foreach($dir in $all) #foreach($sample in ${dir.samples}) #foreach($pair in ${sample.files}) #maketarget($pair.forward) #maketarget($pair.reverse) #end #end #foreach($pair in ${dir.undetermined}) #maketarget($pair.forward) #maketarget($pair.reverse) #end #end clean: rm -f all.count $(addsuffix .count, #foreach($dir in $all) #foreach($sample in ${dir.samples}) #foreach($pair in ${sample.files}) ${pair.forward.md5filename} ${pair.reverse.md5filename} #end #end #foreach($pair in ${dir.undetermined}) ${pair.forward.md5filename} ${pair.reverse.md5filename} #end #end )
transform using jsvelocity:
java -jar dist/jsvelocity.jar \ -d all illumina.json \ illumina.vm > Makefile
ouput: same as above
Pierre
PS: This post was generated using the XSLT stylesheet :"github2html.xsl" and https://github.com/lindenb/jvarkit/wiki/Illuminadir.
Publié par
Pierre Lindenbaum
at
12:56 AM
0
commentaires
![]()
Libellés : bioinformatics, json, make, ngs, tool, velocity, xml, xslt
17 October 2013
Rapid prototyping of a read-only Lims using JSON and Apache Velocity.
In a previous post, I showed how to use the Apache Velocity template engine to format JSON data.
Since that post, I've moved my application to a github repository: https://github.com/lindenb/jsvelocity. The project contains a java-based standalone tool to process the JSON data.
Here is an example: The JSON data:
{
individuals:[
{
name: "Riri",
age: 8,
duck: true
},
{
name: "Fifi",
age: 9,
duck: true
},
{
name: "Loulou",
age: 10,
duck: true
}
]
}.... and the velocity template:#foreach($indi in ${all.individuals})
<h1>${indi['name']}</h1>
Age:${indi.age}<br/>${indi.duck}
#end... with the following command line ... $ java -jar dist/jsvelocity.jar \
-f all test.json \
test.vm... produces the following output ... <h1>Riri</h1> Age:8<br/>true <h1>Fifi</h1> Age:9<br/>true <h1>Loulou</h1> Age:10<br/>true
Today I wrote a web version of the tool using the jetty server. I wanted to quickly write a web interface to display various summaries for our NGS experiments.
My JSON input looks like this:
{
"sequencer":[
{
"name":"HiSeq"
},
{
"name":"MiSeq"
}
],
"run":[ {
"sequencer":"HiSeq",
"flowcell":"C2AVTACXX",
"name":"131010_C2AVTACXX_61",
"date":"2013-10-10",
"comment":"A comment",
"samples":[
{
"name":"CD0001",
"meancov": 10
},
{
"name":"CD0002",
"meancov": 20.0
}
,
{
"name":"CD0003",
"meancov": 30.0
}
]
},
{
"sequencer":"MiSeq",
"flowcell":"C3VATACYY",
"name":"131011_C3VATACYY_62",
"date":"2013-10-11",
"comment":"Another comment",
"samples":[
{
"name":"CD0001",
"meancov": 11
},
{
"name":"CD0006",
"meancov": 21.0
}
,
{
"name":"CD0008",
"meancov": null
}
]
},
{
"sequencer":"MiSeq",
"flowcell":"C4VATACYZ",
"name":"131012_C4VATACYZ_63",
"date":"2013-10-12",
"comment":"Another comment",
"samples":[
{
"name":"CD0010",
"meancov":1,
"comment":"Failed, please, re-sequence"
}
]
}
],
"samples":[
{ "name":"CD0001" },
{ "name":"CD0002" },
{ "name":"CD0003" },
{ "name":"CD0004" },
{ "name":"CD0005" },
{ "name":"CD0006" },
{ "name":"CD0007" },
{ "name":"CD0008" },
{ "name":"CD0009" },
{ "name":"CD0010" }
],
"projects":[
{
"name":"Disease1",
"description": "sequencing Project 1",
"samples":["CD0001","CD0002","CD0006","CD0009"]
},
{
"name":"Disease2",
"description": "sequencing Project 2",
"samples":["CD0002","CD0003","CD0008","CD0009"]
}
]
}
One velocity template is used to browse this 'database': https://github.com/lindenb/jsvelocity/blob/master/src/test/resources/velocity/lims.vm.The server is started like:
java -jar dist/webjsvelocity.jar \
-F lims src/test/resources/json/lims.json \
src/test/resources/velocity/lims.vm
2013-10-17 12:43:35.566:INFO:oejs.Server:main: jetty-9.1.0.M0
2013-10-17 12:43:35.602:INFO:oejs.ServerConnector:main: Started ServerConnector@72dcb6{HTTP/1.1}{0.0.0.0:8080}
(...)
And here is a screenshot of the result:That's it,
Pierre
15 October 2013
GNU parallel for #bioinformatics : my notebook
This notebook about GNU parallel follows Ole Tange’s GNU parallel tutorial ( http://www.gnu.org/software/parallel/parallel_tutorial.html ) but I tried to use some bioinformatics-related examples (align with BWA, Samtools, etc.. ).
That's it,
Pierre
Publié par
Pierre Lindenbaum
at
10:53 AM
0
commentaires
![]()
07 October 2013
Software Environment Management with Modules: my notebook
The following question was recently asked on Biostar "Bioinformatics: how to version control small scripts located all over the server". I suggested to put the scripts in a central repository (under git or whatever ) and to use symbolic links in the workspaces to manage the files. On the other hand, Alex Reynolds suggested to use a Module to deploy versions of a given package.
This tool was also suggested on cited by Lex Nederbragt on twitter:
@yokofakun Yep, module system would allow central place for code/binaries. Load module where/when needed. Loading can be scripted too.
/ Lex Nederbragt (@lexnederbragt) October 5, 2013
I've just played with Modules, here is my (short) experience.
I've got two versions of samtools on my server : 0.1.18 and 0.1.19, I want to easily switch from one version to another.
I created a hidden directory in my home:
mkdir ~/.modulesit contains a directory "samtools" that will contain the two 'modules files' for the two versions of samtools:
mkdir ~/.modules/samtools
The directory '~/.modules' is added to the variable ${MODULESPATH} ( The path that the module command searches when looking for modulefiles )
$ export MODULEPATH=${MODULEPATH}:${HOME}/.modules
Create the Module File
'${HOME}/.modules/samtools/0.1.18' for samtools 0.1.18: This module file add the PATH to samtools0.1.18 and bcftools0.1.18#%Module1.0
proc ModulesHelp { } {
global dotversion
puts stderr "\tSamtools"
}
module-whatis "Samtools 0.1.18"
prepend-path PATH /commun/data/packages/samtools-0.1.18/
prepend-path PATH /commun/data/packages/samtools-0.1.18/bcftools
Create the Module File
'${HOME}/.modules/samtools/0.1.19' for samtools 0.1.19: This module file add the PATH to samtools0.1.19 and bcftools0.1.19#%Module1.0
proc ModulesHelp { } {
global dotversion
puts stderr "\tSamtools"
}
module-whatis "Samtools 0.1.19"
prepend-path PATH /commun/data/packages/samtools-0.1.19/
prepend-path PATH /commun/data/packages/samtools-0.1.19/bcftools
We also create a file '${HOME}/.modules/samtools/.version' to define the default version of samtools:
#%Module1.0 set ModulesVersion "0.1.19"
On startup the shell doesn't know samtools or bcftools:
$ which samtools /usr/bin/which: no samtools in (/commun/sge_root/bin/lx24-amd64:/usr/lib64/qt-3.3/bin:/usr/local/bin:/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/sbin:/home/lindenb/bin) $ which bcftools /usr/bin/which: no bcftools in (/commun/sge_root/bin/lx24-amd64:/usr/lib64/qt-3.3/bin:/usr/local/bin:/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/sbin:/home/lindenb/bin)List the available modules :
$ module avail
------------------------------------------------ /usr/share/Modules/modulefiles ------------------------------------------------
dot module-cvs module-info modules mpich2-x86_64 null use.own
---------------------------------------------------- /home/lindenb/.modules ----------------------------------------------------
samtools/0.1.18 samtools/0.1.19(default)Let's load the default configuration for samtools:$ module load samtools
now, the shell know the PATH to samtools:
$ which samtools /commun/data/packages/samtools-0.1.19/samtools $ which bcftools /commun/data/packages/samtools-0.1.19/bcftools/bcftools
Unload the module for samtools:
$ module unload samtools $ which samtools /usr/bin/which: no samtools in (/commun/sge_root/bin/lx24-amd64:/usr/lib64/qt-3.3/bin:/usr/local/bin:/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/sbin:/home/lindenb/bin) $ which bcftools /usr/bin/which: no bcftools in (/commun/sge_root/bin/lx24-amd64:/usr/lib64/qt-3.3/bin:/usr/local/bin:/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/sbin:/home/lindenb/bin)
Now load the (old) version 0.1.18 of samtools
$ module load samtools/0.1.18
The old versions of samtools and bcftools are now in the $PATH:
$ which samtools /commun/data/packages/samtools-0.1.18/samtools $ which bcftools /commun/data/packages/samtools-0.1.18/bcftools/bcftoolsThat's it,
Pierre
Publié par
Pierre Lindenbaum
at
9:35 PM
1 commentaires
![]()
Libellés : bioinformatics, linux, samtools, tips, tool
26 September 2013
Presentation : File formats for Next Generation Sequencing
Here is my presentation for the course "File formats for Next Generation Sequencing", I gave monday at the University of Nantes.
Publié par
Pierre Lindenbaum
at
9:01 PM
0
commentaires
![]()
Libellés : bam, bioinformatics, format, ngs, vcf
Presentation: Advanced NCBI
Here is my presentation for the course "Advanced NCBI", I gave yesterday at the University of Nantes.
Publié par
Pierre Lindenbaum
at
8:58 PM
0
commentaires
![]()
Libellés : api, bioinformatics, blast, ncbi, presentation
10 September 2013
Building a simple & stupid LIMS with the Eclipse Modeling Framework (#EMF), my notebook.
I played with the Eclipse Modeling Framework (EMF) and created a simple interface to manage a list of sequenced samples.
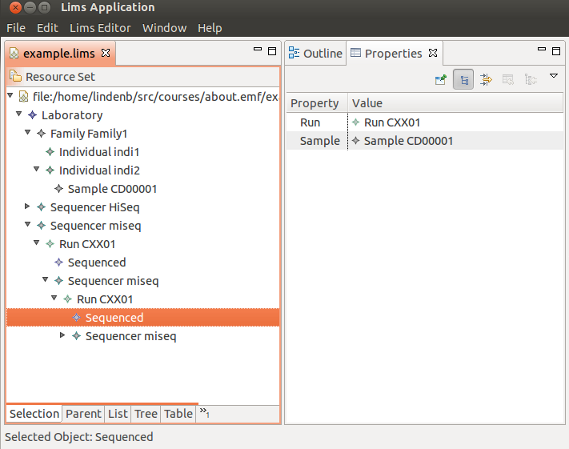
I've uploaded my notes on slideshare:
That's it,
Pierre
26 July 2013
g1kv37 vs hg19
In order to create a class to translate the chromosome names from one naming convention to another. I've compared the MD5 sums of the human genome versions g1k/v37 and ucsc/hg19. Here is the java program to create the MD5s:
The MD5 sums were extracted as follow:
Here are the common chromosomes, joined on the hash-sum:
And here are the unpairable data:
I knew the problem for chrY ( http://www.biostars.org/p/58143/) but not for chr3.. What is the problem for this chromosome ?
Edit: Here are the number of bases for UCSC/chr3:
{T=58760485, G=38670110, A=58713343, C=38653197, N=3225295}and for g1kv37:{T=58760485, G=38670110, A=58713343, R=2, C=38653197, M=1, N=3225292}That's it,
Pierre.
Publié par
Pierre Lindenbaum
at
8:04 PM
2
commentaires
![]()
Libellés : bioinformatics, code, genetics, java
18 July 2013
Running a picard tool in the #KNIME workflow engine
http://www.knime.org/ is "a user-friendly graphical workbench for the entire analysis process: data access, data transformation, initial investigation, powerful predictive analytics, visualisation and reporting". In this post, I'll show how to invoke an external java program, and more precisely a tool from the picard library from with knime. The workflow: load a list of BAM filenames, invoke SortSam and display the names of the sorted files.
Construct the following workflow:
Edit the FileReader node and load a list of paths to the BAMs
Edit the properties of the java node, in the "Additional Libraries" tab, load the jar of SortSam.jar
Edit the java snippet node, create a new column SORTED_BAM for the output.
and copy the following code:
// Your custom imports:
import net.sf.picard.sam.SortSam;
import java.io.File;
----------------------------------------------------------
// Enter your code here:
File input=new File(c_BAM);
//build the output filename
out_SORTED = input.getName();
if(!(out_SORTED.endsWith(".sam") || out_SORTED.endsWith(".bam")))
{
throw new Abort("not a SAM/BAM :"+c_BAM);
}
int dot=out_SORTED.lastIndexOf('.');
out_SORTED=new File(input.getParentFile(),out_SORTED.substring(0, dot)+"_sorted.bam").getPath();
//create a new instance of SortSam
SortSam cmd=new SortSam();
//invoke the instance
int ret=cmd.instanceMain(new String[]{
"I="+input.getPath(),
"O="+out_SORTED,
"SO=coordinate",
"VALIDATION_STRINGENCY=LENIENT",
"CREATE_INDEX=true",
"MAX_RECORDS_IN_RAM=500000"
});
if(ret!=0)
{
throw new Abort("SortSam failed with: "+c_BAM+" "+out_SORTED);
}
Execute KNIME, picard runs the job, and get the names of the sorted BAMs:Edit:
The workflow was uplodaded on MyExperiment at http://www.myexperiment.org/workflows/3654.html.
That's it,
Pierre