Inside Jvarkit: view BAM, cut, stats, head, tail, shuffle, downsample, group-by-gene VCFs...
Here are a few tools I recently wrote (and reinvented) for Jvarkit.
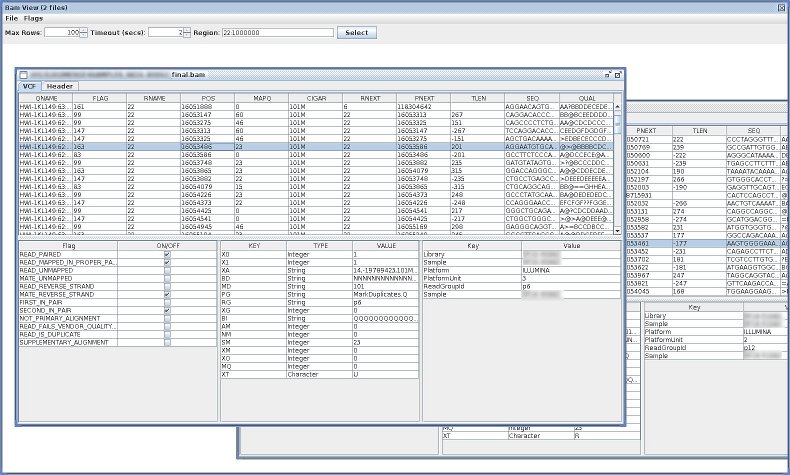
- BamViewGui
- a simple java-Swing-based BAM viewer.
- VcfShuffle
- Shuffle a VCF.
- GroupByGene
- Group VCF data by Gene
$ curl -s -k "https://raw.github.com/arq5x/gemini/master/test/test4.vep.snpeff.vcf" |\ java -jar dist/groupbygene.jar |\ head | column -t #chrom min.POS max.POS gene.name gene.type samples.affected count.variations M10475 M10478 M10500 M128215 chr10 52004315 52004315 ASAH2 snpeff-gene-name 2 1 0 0 1 1 chr10 52004315 52004315 ASAH2 vep-gene-name 2 1 0 0 1 1 chr10 52497529 52497529 ASAH2B snpeff-gene-name 2 1 0 1 1 0 chr10 52497529 52497529 ASAH2B vep-gene-name 2 1 0 1 1 0 chr10 48003992 48003992 ASAH2C snpeff-gene-name 3 1 1 1 1 0 chr10 48003992 48003992 ASAH2C vep-gene-name 3 1 1 1 1 0 chr10 126678092 126678092 CTBP2 snpeff-gene-name 1 1 0 0 0 1 chr10 126678092 126678092 CTBP2 vep-gene-name 1 1 0 0 0 1 chr10 135336656 135369532 CYP2E1 snpeff-gene-name 3 2 0 2 1 1
- DownSampleVcf
- Down sample a VCF.
- VcfHead
- Print the first variants of a VCF.
- VcfTail
- Print the last variants of a VCF
- VcfCutSamples
- Select/Exclude some samples from a VCF
- VcfStats>
- Generate some statistics from a VCF. The ouput is a XML file that can be processed with xslt.
$ curl "https://raw.github.com/arq5x/gemini/master/test/test4.vep.snpeff.vcf" |\
java -jar dist/vcfstats.jar |\
xmllint --format -
<?xml version="1.0" encoding="UTF-8"?>
<vcf-statistics version="314bf88924a4003e6d6189ad3280d8b4df485aa1" input="stdin" date="Thu Dec 12 16:20:14 CET 2013">
<section name="General">
<statistics name="general" description="general">
<counts name="general" description="General" keytype="string">
<property key="num.dictionary.chromosomes">93<
(...)
That's it,
Pierre