A Tiny Genome Browser (XHTML/Javascript/json/svg)
I'm currently working on a small set of NGS data and all I need is a simple genome browser to display my mutations while being able to select some SNPs according to some criteria (is it a known rs## , what is the prediction with SIFT or/and Polyphen ?, etc... ). As it is a relative small amount of data, it can be embedded in a webpage without needing a web server.
First, I need a description of the genes as JSON: I described yesterday how I generated this file using mysql, XSLT and the data from the UCSC:
var knownGenes=[{"name":"uc001aaa.3","chrom":"chr1","strand":"+","txStart":11873,"txEnd":14409,"cdsStart":11873,"cdsEnd":11873,"exonCount":3,"exonStarts":[11873,12612,13220],"exonEnds":[12227,12721,14409],"proteinID":"","alignID":"uc001aaa.3"}
,{"name":"uc010nxq.1","chrom":"chr1","strand":"+","txStart":11873,"txEnd":14409,"cdsStart":12189,"cdsEnd":13639,"exonCount":3,"exonStarts":[11873,12594,13402],"exonEnds":[12227,12721,14409],"proteinID":"B7ZGX9","alignID":"uc010nxq.1"}
,{"name":"uc010nxr.1","chrom":"chr1","strand":"+","txStart":11873,"txEnd":14409,"cdsStart":11873,"cdsEnd":11873,"exonCount":3,"exonStarts":[11873,12645,13220],"exonEnds":[12227,12697,14409],"proteinID":"","alignID":"uc010nxr.1"},
(...)];
,{"name":"uc010nxq.1","chrom":"chr1","strand":"+","txStart":11873,"txEnd":14409,"cdsStart":12189,"cdsEnd":13639,"exonCount":3,"exonStarts":[11873,12594,13402],"exonEnds":[12227,12721,14409],"proteinID":"B7ZGX9","alignID":"uc010nxq.1"}
,{"name":"uc010nxr.1","chrom":"chr1","strand":"+","txStart":11873,"txEnd":14409,"cdsStart":11873,"cdsEnd":11873,"exonCount":3,"exonStarts":[11873,12645,13220],"exonEnds":[12227,12697,14409],"proteinID":"","alignID":"uc010nxr.1"},
(...)];
Second, I need a JSON description of my data:
/** these are ==RANDOM== data */
var variations=[
{"chrom":"chr1","start":12874,"end":12875,"ref":"A","alt":"G","struct":false,"depth":8,"sift":"TOLERATED","pph2div":"benign","pph2var":"benign","rs":null,"hq":true,"id":0},
{"chrom":"chr1","start":792398,"end":792399,"ref":"A","alt":"G","struct":false,"depth":89,"sift":"TOLERATED","pph2div":"probably damaging","pph2var":"benign","rs":null,"hq":true,"id":1},
{"chrom":"chr1","start":959761,"end":959762,"ref":"A","alt":"G","struct":false,"depth":76,"sift":"DAMAGING","pph2div":"probably damaging","pph2var":"benign","rs":null,"hq":true,"id":2}
,(...)];
var variations=[
{"chrom":"chr1","start":12874,"end":12875,"ref":"A","alt":"G","struct":false,"depth":8,"sift":"TOLERATED","pph2div":"benign","pph2var":"benign","rs":null,"hq":true,"id":0},
{"chrom":"chr1","start":792398,"end":792399,"ref":"A","alt":"G","struct":false,"depth":89,"sift":"TOLERATED","pph2div":"probably damaging","pph2var":"benign","rs":null,"hq":true,"id":1},
{"chrom":"chr1","start":959761,"end":959762,"ref":"A","alt":"G","struct":false,"depth":76,"sift":"DAMAGING","pph2div":"probably damaging","pph2var":"benign","rs":null,"hq":true,"id":2}
,(...)];
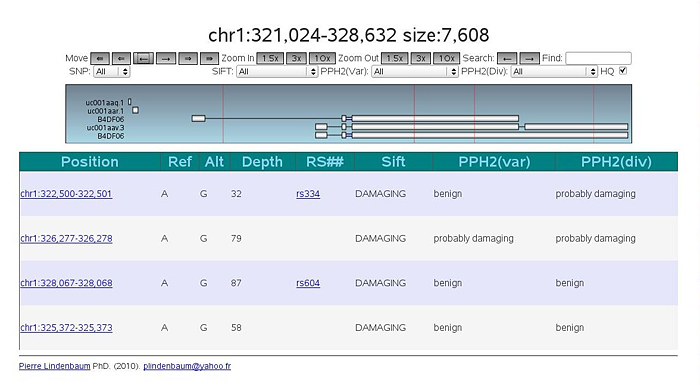
Third, I wrote a small genome browser using javascript , displaying the data as SVG and using the two previous files. Here is a screenshot:
I posted a random sample of the files on http://gist.github.com/508676, feel free to play with it.
That's it
Pierre
4 comments:
You know jbrowse is perfect for this stuff. You just need GFF3 inputs. I use it for this kind of browser all the time.
... but jbrowse requires an external server.
Well, I just use 'python -m SimpleHTTPServer' inside a directory. Works pretty well at serving anything.
Thanks, nice to know
Post a Comment