The EBI/IntAct Web-Service API, my notebook
This post covers my experience with the IntAct API at EBI. IntAct provides a freely available, open source database system and analysis tools for protein interaction data. All interactions are derived from literature curation or direct user submissions and are freely available.
This web service is invoked for searching binary interactions, it is described (but not documented...) as a WSDL file at http://www.ebi.ac.uk/intact/binary-search-ws/binarysearch?wsdl
Glassfih, the Java Application Server from Sun, comes with a tool called wsimport. It generates a set of java files used to handle this web-service from the wsdl file.
Here are the generated java files :
./uk/ac/ebi/intact/binarysearch/wsclient/generated/ObjectFactory.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/FindBinaryInteractionsResponse.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/FindBinaryInteractionsLimitedResponse.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/FindBinaryInteractionsLimited.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/SimplifiedSearchResult.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/GetVersionResponse.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/package-info.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/FindBinaryInteractions.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/GetVersion.java
./uk/ac/ebi/intact/binarysearch/wsclient/generated/BinarySearch.java
AFAIK, the WSDL file contained almost no documentation about this service, but eclipse helped me to find the correct methods thanks to the completion of the code editor.
Here is the short program I just wrote: it connects to the webservice and retrieves all the binary interactions with NSP3
import uk.ac.ebi.intact.binarysearch.wsclient.generated.BinarySearchService;
import uk.ac.ebi.intact.binarysearch.wsclient.generated.SimplifiedSearchResult;
public class IntActClient
{
/**
* @param args
*/
public static void main(String[] args) {
try
{
final String query="NSP3";
BinarySearchService service=new BinarySearchService();
BinarySearch port=service.getBinarySearchPort();
SimplifiedSearchResult ssr= port.findBinaryInteractionsLimited(query, 0,500);
System.out.println("#first-result "+ssr.getFirstResult());
System.out.println("#max-results "+ssr.getMaxResults());
System.out.println("#total-results "+ssr.getTotalResults());
System.out.println("#luceneQuery "+ssr.getLuceneQuery());
for(String line:ssr.getInteractionLines())
{
System.out.println(line);
}
}
catch(Throwable err)
{
err.printStackTrace();
}
}
}
The result:
#max-results 500
#total-results 7
#luceneQuery identifiers:nsp3 pubid:nsp3 pubauth:nsp3 species:nsp3 type:nsp3 detmethod:nsp3 interact
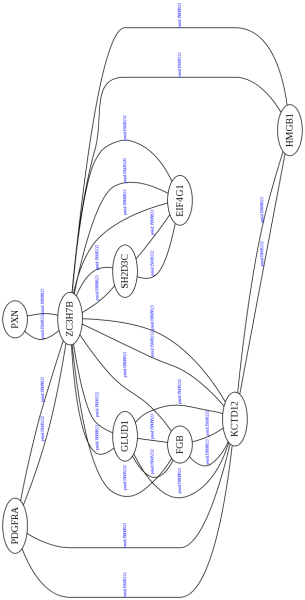
uniprotkb:Q8N5H7|intact:EBI-745980 uniprotkb:O43281|intact:EBI-718488 uniprotkb:SH2D3C uniprotkb:EFS
intact:EBI-1263954 uniprotkb:Q00721|intact:EBI-1263962 - uniprotkb:S7 - uniprotkb:NCVP4|uniprotkb:vn....
uniprotkb:Q00721|intact:EBI-1263962 intact:EBI-1263971 uniprotkb:S7 - uniprotkb:NCVP4|uniprotkb:vn34....
uniprotkb:Q00721|intact:EBI-1263962 uniprotkb:Q00721|intact:EBI-1263962 uniprotkb:S7 uniprotkb:S7 un...
uniprotkb:Q00721|intact:EBI-1263962 uniprotkb:Q9UGR2|intact:EBI-948845 uniprotkb:S7 uniprotkb:ZC3H7B....
uniprotkb:Q04637|intact:EBI-73711 uniprotkb:Q00721|intact:EBI-1263962 uniprotkb:EIF4G1 uniprotkb:S7....
uniprotkb:Q04637|intact:EBI-73711 uniprotkb:P03536|intact:EBI-296448 uniprotkb:EIF4G1 uniprotkb:S7 u...
Ok, it was easy but I'm a little bit disappointed here because the result was 'just' a set of tab delimited lines (and where is the documentation about those columns ??) and I would have rather expected a set of XML objects.
update: the format of the columns was described here:ftp://ftp.ebi.ac.uk/pub/databases/intact/current/psimitab/README.
That's it for tonight....
Pierre