(fig. 4): add one or more organism. (Homo Sapiens already inserted by default)
(fig. 1): add one or more protein.
(fig. 3): add one or more article that will be used as an evidence for an interaction.
(fig. 2): add one or more technology that was used to characterize an interaction.
: add one or more cellular component using Gene Ontology (GO:0005575 \"cellular component\" was inserted by default)
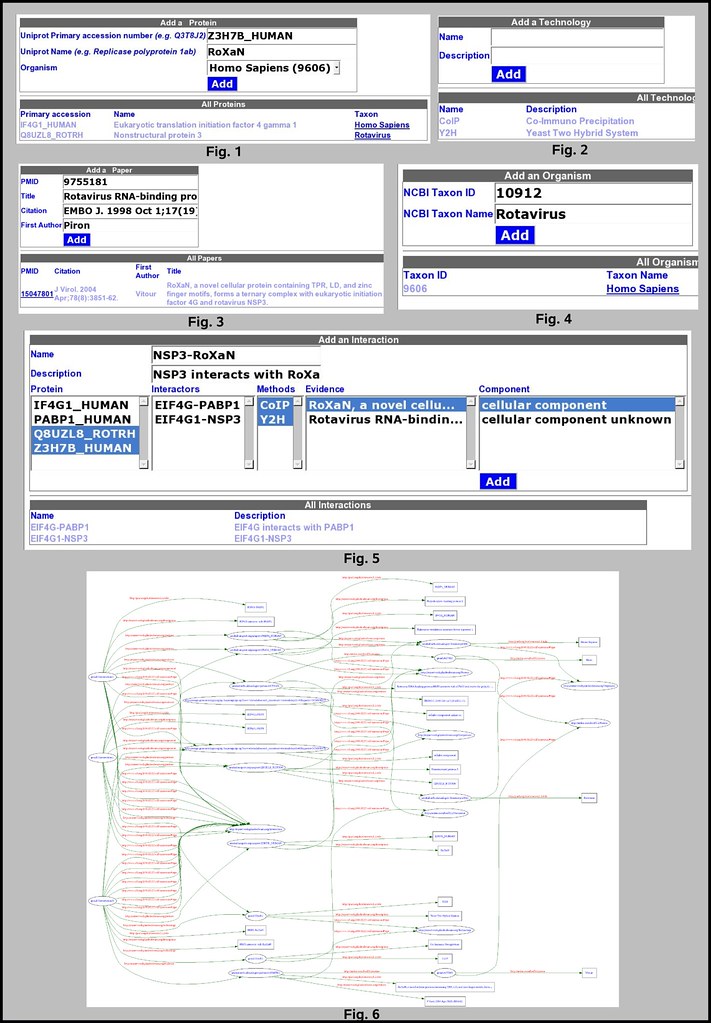
(fig. 5):
(fig. 6): I choose to display the content of the database using
. Such format can then be validated and visualized using the
, etc.... I also used the
as an URI for my resources.
We then create the database if does not exist. The file is created in firefox in ${HOME}/.mozilla/firefox/<profile-id>/Google Gears for Firefox/<server>/mynetwork#database
I create the tables just by invoking some standards SQL 'CREATE TABLE' statements. I also insert some default values (e.g. human organism)
When a data is about to be inserted we check all the fields and we insert them using SQL: INSERT INTO
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "
http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="
http://www.w3.org/1999/xhtml">
<head>
<script type="text/javascript" src="gears_init.js"></script>
<script type="text/javascript" src="network.js"></script>
<link rel="stylesheet" type="text/css" href="./network.css" />
<title>My Biological Network</title>
</head>
<body onload="init()">
<h1>My Biological Network</h1>
<p>Pierre Lindenbaum PhD <a href="
mailto:plindenbaum@yahoo.fr">plindenbaum@yahoo.fr</a><br/><a href="
http://plindenbaum.blogspot.com">
http://plindenbaum.blogspot.com</a><br/><address>Bioinformatics department<br/><a href="
http://www.integragen.com">Integragen S.A.</a><br/>Evry, France</address></p>
<p/>
<div>
<button onclick="javascript:showCard('home-pane');">Home</button>
<button onclick="showOrganismPane()">Organisms</button>
<button onclick="showProteinPane()">Proteins</button>
<button onclick="showPaperPane()">Papers</button>
<button onclick="showTechnologyPane()">Technology</button>
<button onclick="showComponentPane()">Component</button>
<button onclick="showInteractionPane()">Interactions</button>
<button onclick="showRDFPane()">RDF</button>
</div>
<div style="color:red;" id="stderr"></div>
<p/>
<!-- ====================================== ORGANISM ====================================== -->
<div style="display:none;" id="organism-pane">
<table>
<caption>Add an Organism</caption>
<tr><th>NCBI Taxon ID <i>(e.g. 10912)</i></th><td><input id="organism-input-id" length="10"/></td></tr>
<tr><th>NCBI Taxon Name <i>(e.g. Rotavirus)</i></th><td><input id="organism-input-name" length="10"/></td></tr>
<tr><th/><td><button onclick="addOrganism()">Add</button></td></tr>
</table>
<hr/>
<table width="80%">
<caption>All Organisms</caption>
<thead>
<tr><th>Taxon ID</th><th>Taxon Name</th></tr></tr>
</thead>
<tbody id="organism-table">
</tbody>
</table>
</div>
<!-- ====================================== COMPONENT ====================================== -->
<div style="display:none;" id="component-pane">
<table>
<caption>Add a Component</caption>
<tr><th>Name</th><td><input id="component-input-name" length="10"/></td></tr>
<tr><th>GO</th><td><input id="component-input-go" length="10"/></td></tr>
<tr><th/><td><button onclick="addComponent()">Add</button></td></tr>
</table>
<hr/>
<table width="80%">
<caption>All Components</caption>
<thead>
<tr><th>Name</th><th>GO</th></tr>
</thead>
<tbody id="component-table">
</tbody>
</table>
</div>
<!-- ====================================== TECHNOLOGY ====================================== -->
<div style="display:none;" id="technology-pane">
<table>
<caption>Add a Technology</caption>
<tr><th>Name</th><td><input id="technology-input-name" length="50"/></td></tr>
<tr><th>Description</th><td><input id="technology-input-desc" length="50"/></td></tr>
<tr><th/><td><button onclick="addTechnology()">Add</button></td></tr>
</table>
<hr/>
<table width="80%">
<caption>All Technologies</caption>
<thead>
<tr><th>Name</th><th>Description</th></tr></tr>
</thead>
<tbody id="technology-table">
</tbody>
</table>
</div>
<!-- ====================================== PROTEIN ====================================== -->
<div style="display:none;" id="protein-pane">
<table>
<caption>Add a Protein</caption>
<tr><th>Uniprot accession number <i>(e.g. Q3T8J2)</i></th><td><input id="protein-input-acn" length="10"/></td></tr>
<tr><th>Uniprot Name <i>(e.g. Replicase polyprotein 1ab)</i></th><td><input id="protein-input-name" length="10"/></td></tr>
<tr><th>Organism</th><td><select id="protein-input-taxon" length="10"><option>A</option></select></td></tr>
<tr><th/><td><button onclick="addProtein()">Add</button></td></tr>
</table>
<hr/>
<table width="80%">
<caption>All Proteins</caption>
<thead>
<tr><th>Primary accession</th><th>Name</th><th>Taxon</th></tr></tr>
</thead>
<tbody id="protein-table">
</tbody>
</table>
</div>
<!-- ====================================== PAPER ====================================== -->
<div style="display:none;" id="paper-pane">
<table>
<caption>Add a Paper</caption>
<tr><th>PMID</th><td><input id="paper-input-pmid" length="10"/></td></tr>
<tr><th>Title</th><td><input id="paper-input-title" length="50"/></td></tr>
<tr><th>Citation</th><td><input id="paper-input-citation" length="50"/></td></tr>
<tr><th>First Author</th><td><input id="paper-input-author" length="50"/></td></tr>
<tr><th/><td><button onclick="addPaper()">Add</button></td></tr>
</table>
<hr/>
<table width="80%">
<caption>All Papers</caption>
<thead>
<tr><th>PMID</th><th>Citation</th><th>First Author</th><th>Title</th></tr></tr>
</thead>
<tbody id="paper-table">
</tbody>
</table>
</div>
<!-- ====================================== INTERACTION ====================================== -->
<div style="display:none;" id="interaction-pane">
<table>
<caption>Add an Interaction</caption>
<tr><th>Name</th><td colspan="4"><input id="interaction-input-name" length="50"/></td></tr>
<tr><th>Description</th><td colspan="4"><input id="interaction-input-desc" length="50"/></td></tr>
<tr>
<th>Protein</th>
<th>Interactors</th>
<th>Methods</th>
<th>Evidences</th>
<th>Components</th></tr>
<tr>
<td><select id="interactors-input-proteins" size="5" multiple="true"/></td>
<td><select id="interactors-input-interactors" size="5" multiple="true"></td>
<td><select id="interactors-input-technologies" size="5" multiple="true"></td>
<td><select id="interactors-input-evidences" size="5" multiple="true"></td>
<td><select id="interactors-input-components" size="5" multiple="true"></td>
</tr>
<tr><th colspan="4"/><td><button onclick="addInteraction()">Add</button></td></tr>
</table>
<hr/>
<table width="80%">
<caption>All Interactions</caption>
<thead>
<tr><th>Name</th><th>Description</th></tr></tr>
</thead>
<tbody id="interaction-table">
</tbody>
</table>
</div>
<!-- ====================================== RDF ====================================== -->
<div style="display:none;" id="rdf-pane">
<h2>RDF Pane</h2>
<textarea wrap="off" id="rdf-area" rows="20" cols="80"></textarea>
</div>
<!-- ====================================== HOME ====================================== -->
<div style="display:none;" id="home-pane">
<h3>About My Biological Network</h3>
<p><a href="
http://gears.google.com/">Google gears</a> is an open source browser extension that enables web applications to provide offline functionality. The data are stored locally in a fully-searchable relational database using the <a href="
http://www.sqlite.org/">sqlite engine</a>.</p>
<p><b>My Biological Network</b> is a tool I created as a test to play with Google gears: it is used to build a network of protein-protein interactions. It uses Google Gears to record your entries on the <u>local disk</u>, so Gears needs to be installed on your computer. </p>
<p>
Open the tab <b>Organism</b>: add one or more organism. (Homo Sapiens already inserted by default)<br/>
Open the tab <b>Protein</b>: add one or more protein.<br/>
Open the tab <b>Paper</b>: add one or more article that will be used as an evidence for an interaction.<br/>
Open the tab <b>Technology</b>: add one or more technology that was used to characterize an interaction.<br/>
Open the tab <b>Component</b>: add one or more cellular component using Gene Ontology (GO:0005575 \"cellular component\" was inserted by default)<br/>
Open the tab <b>Interaction</b>:<ul>
<li>Name and describe this interaction</li>
<li>Select one or more protein and/or one or more previously defined proteic complex. You <i>Cannot</i> describe self interactions with this tool.<li>
<li>(optional) choose one or more paper/technology/component...</li>
</ul><br/>
Open the <b>RDF table</b>: I choose to display the content of the database using <a href="
http://www.w3.org/RDF/">RDF</a>. Such format can then be validated and visualized using the <a href="
http://www.w3.org/RDF/Validator/">W3C RDF validator</a>, or transformed using <a href="
http://www.w3.org/TR/xslt">XSLT</a>, etc.... I also used the <a href="
http://lsid.sourceforge.net/">life science identifier (LSID)</a> as an URI for my resources.<br/>
</p>
<p>On my computer, the database is stored in <code>$HOME/.mozilla/firefox/<profile-id>/Google Gears for Firefox/<host>/mynetwork#database</code>. The database can be manualy accessed using <a href="
http://www.sqlite.org/">sqlite3</a>:<pre style='color:black;border:1pt solid;background:lightgray;'>sqlite3 mynetwork#database
SQLite version 3.4.0
Enter '.help' for instructions
sqlite> .tables
component interactionhash paper technology
interaction organism prote
sqlite> .schema organism
CREATE TABLE organism(id integer primary key ,name varchar(50) not null unique);
sqlite> select * from organism;
9606|Homo Sapiens
sqlite></pre>
</p>
</div>
<!-- google analytics -->
<script src="
http://www.google-analytics.com/urchin.js"
type="text/javascript">
</script>
<script type="text/javascript">
_uacct = "XXXXXX";
urchinTracker();
</script>
<!-- google analytics -->
</body>
</html>
 X:Map is a genome browser (http://xmap.picr.man.ac.uk/) which uses the google map API and the data from Ensembl. The result is really neat.
X:Map is a genome browser (http://xmap.picr.man.ac.uk/) which uses the google map API and the data from Ensembl. The result is really neat.