Jan Aerts is also writing pARP, a circular genome browser using Ruby and ruby-processing:
My data are stored in big database and it might take some time before all the data are processed and displayed. So my idea was to call the server with some asynchronous ajax queries, retrieve the chunks of data and display each chunk as soon it is returned by the server as soon as it is available.
The code below is a proof of concept. This code is ugly, I wouldn't code things like this for a real piece of software. As a source of data I've used the snp129 and the knownGene tables of the UCSC stored in a mysql database. The server was implemented using PHP.
Client Side
When the document is loaded, the<canvas> element is resized. A first AJAX query is sent to retrieve an array of density of the SNPs on the human chromosome 1. The JSON response is processed, the maximum number of SNPs is found and each item of this array is displayed on the canvas. After that, a second AJAX query is sent to retrieve the density of the genes.<html xmlns="http://www.w3.org/1999/xhtml"><head>
<meta http-equiv="Content-Type" content="text/html; charset=iso-8859-1"/>
<script><![CDATA[
/** the canvas element */
var canvas = null;
/** radius of the canvas */
var radius=500;
/** AJAX request */
var httpRequest=null;
/** Graphics context */
var g=null;
/** length of chrom1 */
var CHR1_LENGTH =248000000.0;
/** window length (pb) */
var windowLength=0;
/** first track is snp129 */
var database="snp129";
/** ajax callback */
function paintSnps()
{
if (httpRequest.readyState == 4) {
// everything is good, the response is received
if (httpRequest.status == 200)
{
var jsondata=eval("("+httpRequest.responseText+")");
var counts=jsondata.counts;
//get the maximum of item
var max=0;
for(var i=0;i< counts.length;++i)
{
if(counts[i].count > max) max= counts[i].count*1.0;
}
var r1= radius/2.0;
if(database=="knownGene")
{
r1+= 2+radius/4.0;
}
//loop over the items
for(var i=0;i< counts.length;++i)
{
var a1= Math.PI*2.0*i/(1.0*counts.length);
var a2= Math.PI*2.0*(i+1)/(1.0*counts.length);
var r2= r1+(counts[i].count/max)*(radius/4.0);
//draw the item
g.beginPath();
g.moveTo( radius + Math.cos(a1)*r1, radius + Math.sin(a1)*r1);
g.lineTo( radius + Math.cos(a1)*r2, radius + Math.sin(a1)*r2);
g.lineTo( radius + Math.cos(a2)*r2, radius + Math.sin(a2)*r2);
g.lineTo( radius + Math.cos(a2)*r1, radius + Math.sin(a2)*r1);
g.stroke();
g.fill();
}
//if it was snp, then look for knownGene, change the coors
if(database=="snp129")
{
database="knownGene";
g.fillStyle = "yellow";
g.strokeStyle = "blue";
setTimeout("fetchDB()",100);
}
}
else
{
//boum!!
}
}
else {
// still not ready
}
}
/** calls the AJAX request */
function fetchDB()
{
httpRequest= new XMLHttpRequest();
httpRequest.onreadystatechange = paintSnps;
httpRequest.open('GET', 'ucsc.php', true);
httpRequest.send("length="+windowLength+"database="+database);
}
/** init document */
function init()
{
canvas=document.getElementById("genome");
//resize canvas
canvas.setAttribute("width",2*radius);
canvas.setAttribute("height",2*radius);
if (!canvas.getContext) return;
g = canvas.getContext('2d');
//paint background
var lineargradient = g.createLinearGradient(radius,0,radius,2*radius);
lineargradient.addColorStop(0,'white');
lineargradient.addColorStop(1,'black');
g.fillStyle = lineargradient;
g.fillRect(0,0,2*radius,2*radius);
g.strokeStyle = "black";
g.strokeRect(0,0,2*radius,2*radius);
g.fillStyle = "red";
g.strokeStyle = "green";
var perimeter= 2*Math.PI*(radius/2.0);
windowLength = Math.round(CHR1_LENGTH/perimeter);
//launch the first ajax request
setTimeout("fetchDB()",100);
}
]]></script>
</head><body onload="init();">
<canvas id="genome" />
</body></html>
<meta http-equiv="Content-Type" content="text/html; charset=iso-8859-1"/>
<script><![CDATA[
/** the canvas element */
var canvas = null;
/** radius of the canvas */
var radius=500;
/** AJAX request */
var httpRequest=null;
/** Graphics context */
var g=null;
/** length of chrom1 */
var CHR1_LENGTH =248000000.0;
/** window length (pb) */
var windowLength=0;
/** first track is snp129 */
var database="snp129";
/** ajax callback */
function paintSnps()
{
if (httpRequest.readyState == 4) {
// everything is good, the response is received
if (httpRequest.status == 200)
{
var jsondata=eval("("+httpRequest.responseText+")");
var counts=jsondata.counts;
//get the maximum of item
var max=0;
for(var i=0;i< counts.length;++i)
{
if(counts[i].count > max) max= counts[i].count*1.0;
}
var r1= radius/2.0;
if(database=="knownGene")
{
r1+= 2+radius/4.0;
}
//loop over the items
for(var i=0;i< counts.length;++i)
{
var a1= Math.PI*2.0*i/(1.0*counts.length);
var a2= Math.PI*2.0*(i+1)/(1.0*counts.length);
var r2= r1+(counts[i].count/max)*(radius/4.0);
//draw the item
g.beginPath();
g.moveTo( radius + Math.cos(a1)*r1, radius + Math.sin(a1)*r1);
g.lineTo( radius + Math.cos(a1)*r2, radius + Math.sin(a1)*r2);
g.lineTo( radius + Math.cos(a2)*r2, radius + Math.sin(a2)*r2);
g.lineTo( radius + Math.cos(a2)*r1, radius + Math.sin(a2)*r1);
g.stroke();
g.fill();
}
//if it was snp, then look for knownGene, change the coors
if(database=="snp129")
{
database="knownGene";
g.fillStyle = "yellow";
g.strokeStyle = "blue";
setTimeout("fetchDB()",100);
}
}
else
{
//boum!!
}
}
else {
// still not ready
}
}
/** calls the AJAX request */
function fetchDB()
{
httpRequest= new XMLHttpRequest();
httpRequest.onreadystatechange = paintSnps;
httpRequest.open('GET', 'ucsc.php', true);
httpRequest.send("length="+windowLength+"database="+database);
}
/** init document */
function init()
{
canvas=document.getElementById("genome");
//resize canvas
canvas.setAttribute("width",2*radius);
canvas.setAttribute("height",2*radius);
if (!canvas.getContext) return;
g = canvas.getContext('2d');
//paint background
var lineargradient = g.createLinearGradient(radius,0,radius,2*radius);
lineargradient.addColorStop(0,'white');
lineargradient.addColorStop(1,'black');
g.fillStyle = lineargradient;
g.fillRect(0,0,2*radius,2*radius);
g.strokeStyle = "black";
g.strokeRect(0,0,2*radius,2*radius);
g.fillStyle = "red";
g.strokeStyle = "green";
var perimeter= 2*Math.PI*(radius/2.0);
windowLength = Math.round(CHR1_LENGTH/perimeter);
//launch the first ajax request
setTimeout("fetchDB()",100);
}
]]></script>
</head><body onload="init();">
<canvas id="genome" />
</body></html>
The server
The (ugly) PHP page is a simple script returning the density of the objects mapped on the chromosome 1 for a given table.<?php
$con=NULL;
function cleanup()
{
if($con!=NULL) mysql_close($con);
flush;
exit;
}
header('Cache-Control: no-cache, must-revalidate');
header('Content-type: application/json');
header("Content-Disposition: attachment; filename=\"result.json\"");
header('Content-type: text/plain');
$con = mysql_connect('localhost', 'anonymous', '');
if (!$con) {
echo "{status:'Error',message:'". mysql_error()."'}";
cleanup();
}
if(!mysql_select_db('hg18', $con))
{
echo "{status:'Error',message:'cannot select db'}";
cleanup();
}
$database="snp129";
if(isset($_GET["database"]))
{
$database=$_GET["database"];
}
$length=1E6;
if(isset($_GET["length"]))
{
$length= (int)$_GET["length"];
}
if($length<=0) $length=1E6;
$nameStart="chromStart";
if($database=="knownGene")
{
$nameStart="txStart";
}
$sql="SELECT CAST(ROUND(".$nameStart."/".$length.") AS SIGNED INTEGER )*".$length.",count(*) from ".$database." where ".
" chrom=\"chr1\" ".
" group by CAST(ROUND(".$nameStart."/".$length.") AS SIGNED INTEGER )*".$length.
" order by 1"
;
$result = mysql_query($sql ,$con );
if(!$result)
{
echo "{status:'Error',message:'".mysql_error($con) ."'}";
cleanup();
}
$found=FALSE;
echo "{status:'OK',";
echo "length:".$length.",";
echo "counts:[";
while ($row = mysql_fetch_array($result))
{
if($found) echo ",\n";
$found=TRUE;
echo "{chromStart:".$row[0].",count:".$row[1]."}";
}
echo "]}";
cleanup();
?>
And here is the kind of JSON document returned by the server:$con=NULL;
function cleanup()
{
if($con!=NULL) mysql_close($con);
flush;
exit;
}
header('Cache-Control: no-cache, must-revalidate');
header('Content-type: application/json');
header("Content-Disposition: attachment; filename=\"result.json\"");
header('Content-type: text/plain');
$con = mysql_connect('localhost', 'anonymous', '');
if (!$con) {
echo "{status:'Error',message:'". mysql_error()."'}";
cleanup();
}
if(!mysql_select_db('hg18', $con))
{
echo "{status:'Error',message:'cannot select db'}";
cleanup();
}
$database="snp129";
if(isset($_GET["database"]))
{
$database=$_GET["database"];
}
$length=1E6;
if(isset($_GET["length"]))
{
$length= (int)$_GET["length"];
}
if($length<=0) $length=1E6;
$nameStart="chromStart";
if($database=="knownGene")
{
$nameStart="txStart";
}
$sql="SELECT CAST(ROUND(".$nameStart."/".$length.") AS SIGNED INTEGER )*".$length.",count(*) from ".$database." where ".
" chrom=\"chr1\" ".
" group by CAST(ROUND(".$nameStart."/".$length.") AS SIGNED INTEGER )*".$length.
" order by 1"
;
$result = mysql_query($sql ,$con );
if(!$result)
{
echo "{status:'Error',message:'".mysql_error($con) ."'}";
cleanup();
}
$found=FALSE;
echo "{status:'OK',";
echo "length:".$length.",";
echo "counts:[";
while ($row = mysql_fetch_array($result))
{
if($found) echo ",\n";
$found=TRUE;
echo "{chromStart:".$row[0].",count:".$row[1]."}";
}
echo "]}";
cleanup();
?>
{status:'OK',
length:1000000,
counts:[
{chromStart:0,count:6191},
{chromStart:1000000,count:8897},
{chromStart:2000000,count:5559},
{chromStart:3000000,count:6671},
{chromStart:4000000,count:6398},
{chromStart:5000000,count:5462},
{chromStart:6000000,count:5678},
{chromStart:7000000,count:4737},
{chromStart:8000000,count:5313},
{chromStart:9000000,count:5148},
{chromStart:10000000,count:4055},
{chromStart:11000000,count:5012},
{chromStart:12000000,count:5363},
{chromStart:13000000,count:10165},
(...)
{chromStart:239000000,count:5502},
{chromStart:240000000,count:6173},
{chromStart:241000000,count:7928},
{chromStart:242000000,count:3800},
{chromStart:243000000,count:5503},
{chromStart:244000000,count:7120},
{chromStart:245000000,count:6148},
{chromStart:246000000,count:6015},
{chromStart:247000000,count:5337}
]
}
length:1000000,
counts:[
{chromStart:0,count:6191},
{chromStart:1000000,count:8897},
{chromStart:2000000,count:5559},
{chromStart:3000000,count:6671},
{chromStart:4000000,count:6398},
{chromStart:5000000,count:5462},
{chromStart:6000000,count:5678},
{chromStart:7000000,count:4737},
{chromStart:8000000,count:5313},
{chromStart:9000000,count:5148},
{chromStart:10000000,count:4055},
{chromStart:11000000,count:5012},
{chromStart:12000000,count:5363},
{chromStart:13000000,count:10165},
(...)
{chromStart:239000000,count:5502},
{chromStart:240000000,count:6173},
{chromStart:241000000,count:7928},
{chromStart:242000000,count:3800},
{chromStart:243000000,count:5503},
{chromStart:244000000,count:7120},
{chromStart:245000000,count:6148},
{chromStart:246000000,count:6015},
{chromStart:247000000,count:5337}
]
}
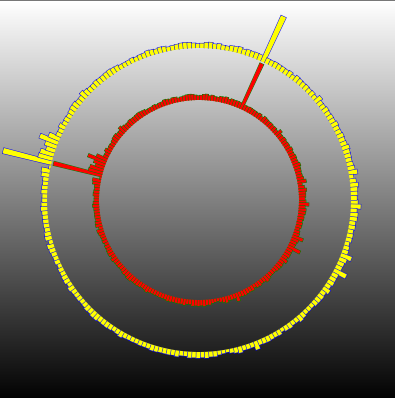
Result
That's it
PS: Hum, yes I know , it's not as fast/beautiful as GenoDive that was introduced at Biohackathon.
Pierre


Hey Pierre,
ReplyDeleteThe approach I am trying to take with pARP is to build an index beforehand that contains information at different levels. I will create a first aggregate level with bins of e.g. 100 bp. So each bin has a start and a stop (i.c. 1-100, 101-200,...), as well as some aggregate values (e.g. SNP count of child bins). Then I'd create the next level, taking 100 of those bins and putting those together. Again, this bin at the next level has a start, stop and SNP count. If we do that for the whole genome, it's lightning fast (a) to get all features from a small region, and (b) to draw pictures. After all, if you have a circle with radius 100, you only have 2*PI*100=628 pixels you can use. It doesn't make sense to try and draw 20,000,000 SNPs at that point. What you can do instead is search that level in the index tree that has about 628 bins in it. Then you just have to draw 628 values instead of 20,000,000.
Nice work, by the way :-)
ReplyDelete